|
Statistic Analysis and Web Figure 1:
This PDF file contains SUPPLEMENTARY APPENDIX I to the paper, which provides a detailed description of the statistical methods used. This file also includes Web Figure 1. Gene-Expression Signature Defined by Hierarchical Clustering.
Patient Data:
This text file contains clinical data from the 240 patients used to formulate and test the gene expression-based outcome predictor. Samples from these patients were divided into a Training (Preliminary) set and a Validation set for statistical analysis. The gene expression signature averages used to compute the outcome predictor score for each patient are given as is the outcome predictor score.
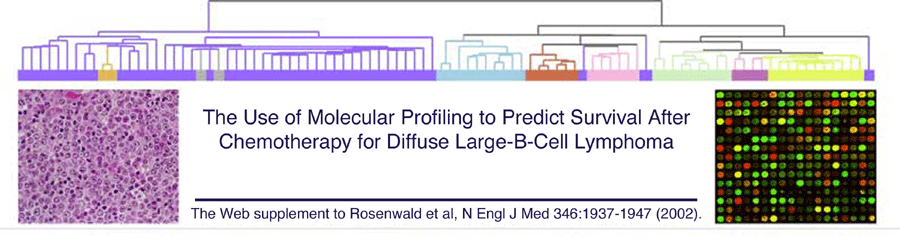
Fig 1A cdt file:
This text file is in the cdt file format for Michael Eisen's Cluster program (http://www.eisenlab.org/software.html). It contains data for Fig 1A in the paper.
- GID is an internal ID for Cluster program.
- UNIQID is a unique ID for Lymphochip microarray feature.
- NAME is a multi-field description of the Lymphochip microarray feature, with each field separated by a vertical bar.
Fields may be empty depending on the information that is available concerning the sequence identity of the microarray feature.
The first field is the Genbank Accession number for a named gene that the microarray feature represents.
The second field is the Genbank Accession number for an IMAGE consortium cDNA clone. For some Lymphochip features, this Genbank accession is derived from the IMAGE cDNA clone that is used to generate the microarray feature and that has been sequence verified (designated by *). For other features, a representative Genbank accession was chosen that matches the sequence of the cDNA clone used to generate the microarray feature, but is not actually the accession associated with that clone (designated by ~). The third field is the Unigene cluster ID for the gene or cDNA clone represented by the microarray feature. The fourth field is the Unigene cluster title. Lymphochip microarray clones that have not been sequence verified are designated by their Lymphochip unique ID in this field.
- GWEIGHT is an internal parameter for Cluster program.
- Array columns contain measurement of the log2 CY5/CY3 expression ratio for each feature. The CY5 channel represents the experimental sample mRNA. The CY3 channel represents the control reference mRNA pool used in every Lymphocip microarray experiment.
Web Figure 1 Data file:
This text file contains data for Web Figure 1 (See Supplement Data).
- UNIQID is a unique ID for Lymphochip microarray feature.
- NAME is a multi-field description of the Lymphochip microarray feature, with each field separated by a vertical bar. See Fig 1A cdt file description above for details.
- Array columns contain measurements of the log2 CY5/CY3 expression ratio for each feature. The CY5 channel represents the experimental sample mRNA. The CY3 channel represents the control reference mRNA pool used in every Lymphochip microarray experiment.
Web Figure 1 Intensity files:
These 2 text files contain CY3 and CY5 intensity data for Web Figure 1 (See Supplement Data file). The measurements represent the mean of spot intensity minus the medium of the background intensity, as calculated by the Genepix software package (Axon).
- UNIQID is a unique ID for Lymphochip microarray feature.
- Array columns contain intensity measurement for each feature. The CY5 channel represents the experimental sample mRNA. The CY3 channel represents the control reference mRNA pool used in every Lymphochip microarray experiment.
Gene Expression Signature files:
These text files contain lists of Lymphochip microarray features which were considered to be gene expression signatures of different biological processes.
- UNIQID is a unique ID for a Lymphochip microarray feature.
-
NAME is a multi-field descriptionof the Lymphochip microarray feature, with each field separated by a vertical bar. See the Fig 1A cdt file description above for details.
For questions and/or comments on this website, please e-mail to LLMPP-support.
|