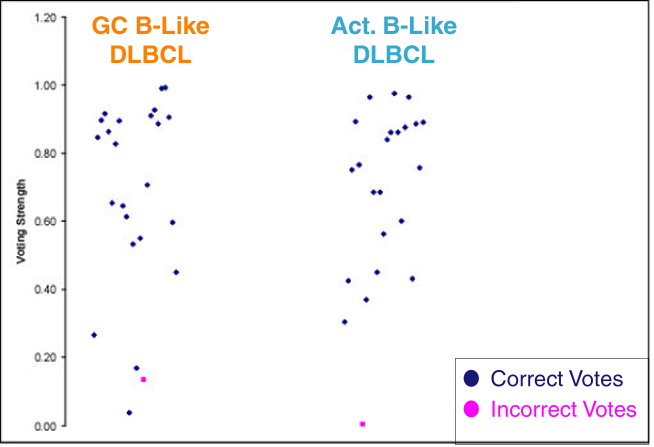
Voting, as per Golub et al. (1999), to distinguish GC B-Like DLBCL from Activated B-Like DLBCL demonstrates the robustness of this disease definition.
For each example, a dataset was generated with that sample deleted. 50 genes were identified that best discriminated between GC B-Like and Activated B-Like DLBCL (the 25 most positively correlated with each disease, using the Golub et al. group distinction score). A vote was taken using these 50 genes, again as described in Golub et al., and a "blind" prediction made for each sample. Only two samples were incorrectly classified, and only a total of 5 had Voting Strengths below the 0.3 cutoff suggested by Golub et al.