|
GeneExplorer is a web application written by
Christian Rees
that allows the exploration of large bodies of gene expression data
generated by DNA microarray experiments using a web browser
as its interface. Gene Explorer is based on the Windows program
TreeView
written by Michael Eisen.
A hierarchical clustering algorithm (Eisen et al.
(1998)) was used to group genes based on similarity in the pattern with which their expression
varied over all samples. The same clustering method was used to group
tumor and cell samples based on similiarities in their expression of these
genes. The data are shown in a matrix format, with each row representing
all the hybridization results for a single cDNA element of the array, and
each column representing the measured expression levels for all genes in a
single sample. To visualize the results, the expression level of each gene
(relative to its median expression level across all samples) was
represented by a color, with red representing expression greater than the
mean, green representing expression less than the mean, and the color
intensity representing the magnitude of the deviation from the mean.
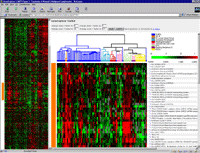 A "bird's eye view" of the data is displayed in a "thumbnail" or
"Radar" pane on the
left, and a blown up view in the "Zoom" pane on the right.
The "Radar" gives an
overview of the general patterns observable in the expression data after it has been submitted to a clustering procedure.
A "bird's eye view" of the data is displayed in a "thumbnail" or
"Radar" pane on the
left, and a blown up view in the "Zoom" pane on the right.
The "Radar" gives an
overview of the general patterns observable in the expression data after it has been submitted to a clustering procedure.
By clicking on a region of interest in the "Radar" view, a regional enlargement will be displayed in the "Zoom" view.
Here an identifier for each clone is visible. For some clones, this identifier refers to a known
human gene. For other clones, the identifier may reflect sequence homology,
but not identity, to a gene as judged by BLAST analysis (labeled "Similar
to ...). The remainder of the clones are listed as "Unknown". If these
"Unknown" clones have been assigned to a Unigene cluster, as defined by
NCBI, the identifier includes "UG Hs." followed by the Unigene
identification number. The identifier is prefaced by an asterisk if the
sequence assignment of the clone has been verified; otherwise the
identifier is in parentheses. Each row also includes links to other
information about the clone.
|











